 Introduction
Introduction
cy2sbml is an open source Cytoscape 2 plugin for the Systems Biology Markup Language SBML.
Latest release
![]() Download
Download
Installation instructions & source code
https://github.com/matthiaskoenig/cy2sbml/
Support & Forum: https://groups.google.com/forum/#!forum/cysbml-cyfluxviz
Bug Tracker: https://github.com/matthiaskoenig/cy2sbml/issues
For Cytoscape 3 use cy3sbml.
Features
cy2sbml provides advanced functionality for the import and work with models encoded in SBML, amongst others the visualization of SBML network annotations within the network context, direct import of models from repositories like biomodels and one-click access to annotation resources and SBML model information and SBML validation.
- Java based SBML parser for Cytoscape based on the java SBML library JSBML
- simple access to SBML models and SBML annotations via BioModels and MIRIAM web services
- supports all versions of SBML
- support of SBML Layout Package
- support of SBML Qualitative Models Package
- provides SBML validation (SBML warnings and errors accessible)
- creates standard network layout based on the SBML species/reaction model
- provides access to RDF based annotation information within the network context
- Navigation menu based on the SBML structure linked to layout and annotation information
- tested with all SBML.org and Biomodels.org test cases (sbml-test-cases-2.0.2, BioModelsDatabase-r21-sbmlfiles)
Citation
Matthias König, Andreas Dräger and Hermann-Georg Holzhütter
CySBML: a Cytoscape plugin for SBML
Bioinformatics. 2012 Jul 5. PMID:22772946
cy2sbml was developed by Matthias König in cooperation with Andreas Dräger from the Center of Bioinformatics Tuebingen (ZBIT) within the Virtual Liver Network.
We thank Camille Laibe for implementing additional BioModel WebService functionality, the Qual Team and Finja Büchel, Florian Mittag, and Nicolas Rodriguez (Qual implementation in JSBML), Sebastian Fröhlich and Clemens Wrzodek (Layout support in JSBML).
Usage guide
Tutorial
The cy2sbml tutorial covers
- Installation
- cy2sbml interface
- import of SBML models
- Access to annotation information
- SBML validation in cy2sbml
- programmatic Interaction with cy2sbml
Menu Bar
The main functionality of CySBML is accessible via the Cytoscape menu bar in the top region of the Cytoscape window

 |
SBML Import | Load SBML files via the File Import Dialog. To import multiple files select multiple files. |
 |
BioModel Import | SBML files from BioModels are loaded via the BioModel Import Dialog. |
 |
SBML Validation | Imported SBML files can be validated. Select the SBML network to validate and click the validation icon. |
 |
Hide/Show CySBML Panel | Changes the visibility of the CySBML Panel for better overview. Initially the CySBML Navigation Panel is hidden and only opened after loading of SBML models. |
 |
Help | CySBML tutorial and help system. |
BioModel Import
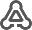 BioModels can be searched via name, person, publication (abstract or pubmed id),
ChEBI (id or name) and UniProt (id or name). The selected models form the result list can than be imported.
BioModels can be searched via name, person, publication (abstract or pubmed id),
ChEBI (id or name) and UniProt (id or name). The selected models form the result list can than be imported.
To load, for instance, all models related to the name glycolysis search by name "glycolysis" select all found models and click "Load Selected". Information for the search results is displayed, selected models are marked in green.
Alternatively models can be imported based on given BioModel identifiers or text containing BioModel identifiers.
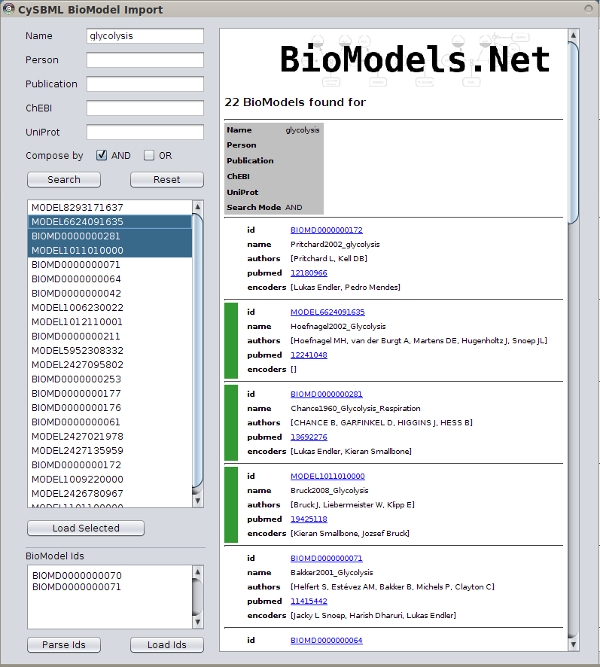
Integration with Cytoscape and Other Plugins
cysbml integrates seamlessly with the Cytoscape ecosystem consisting of the Cytoscape core and plugins. Many plugins work out of the box with Cytoscape, thereby providing additional functionality for SBML files. For instance NetworkAnalyzer to calculate topological parameters of the models, NetMatch to find network motifs within SBML models or cy2fluxviz to visualize flux distributions in SBML models.
cysbml provides the SBML information as Cytoscape node and edge attributes under well defined names given in the CySBMLConstants class. For in detailed information of interaction with CySBML see the tutorial example.
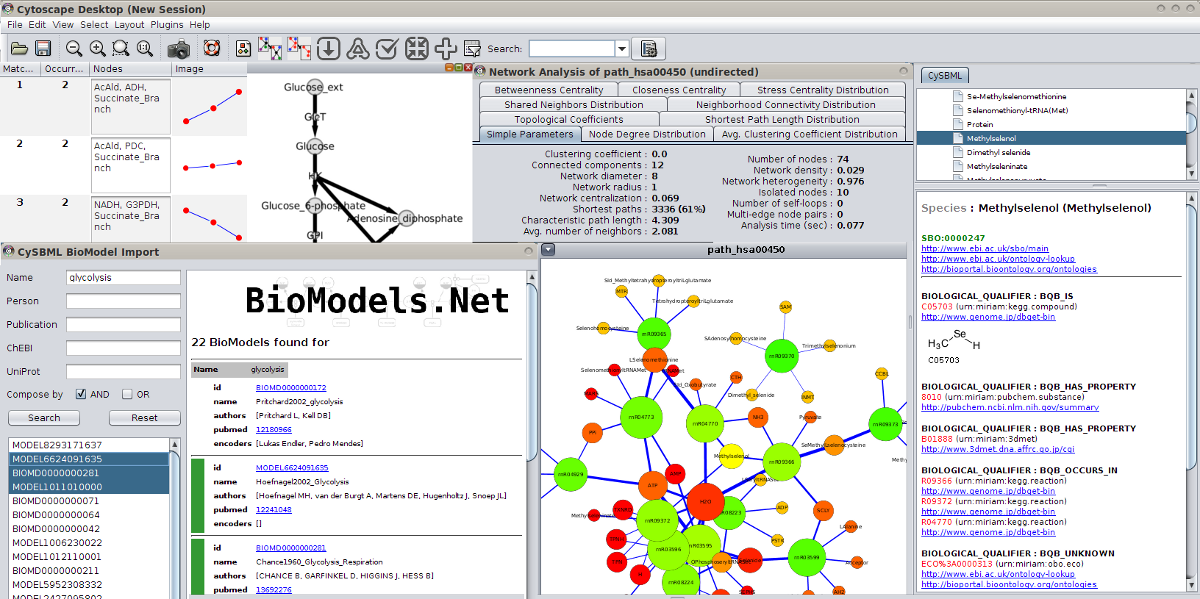
SBML Annotations
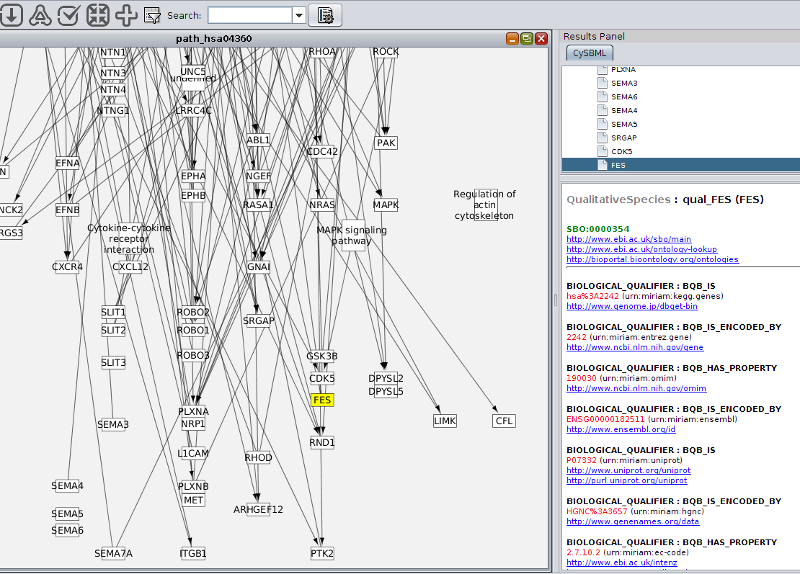
MIRIAM and SBO annotations are accessible via the CySBML Navigation Panel in the Cytoscape Results Panel.
Annotation information is loaded on the fly for selected nodes in the network, like in the example the information for glucose-6 phosphate for the BioModel of Pritchard2002. Crosslinks to additional resources are opened in the browser.
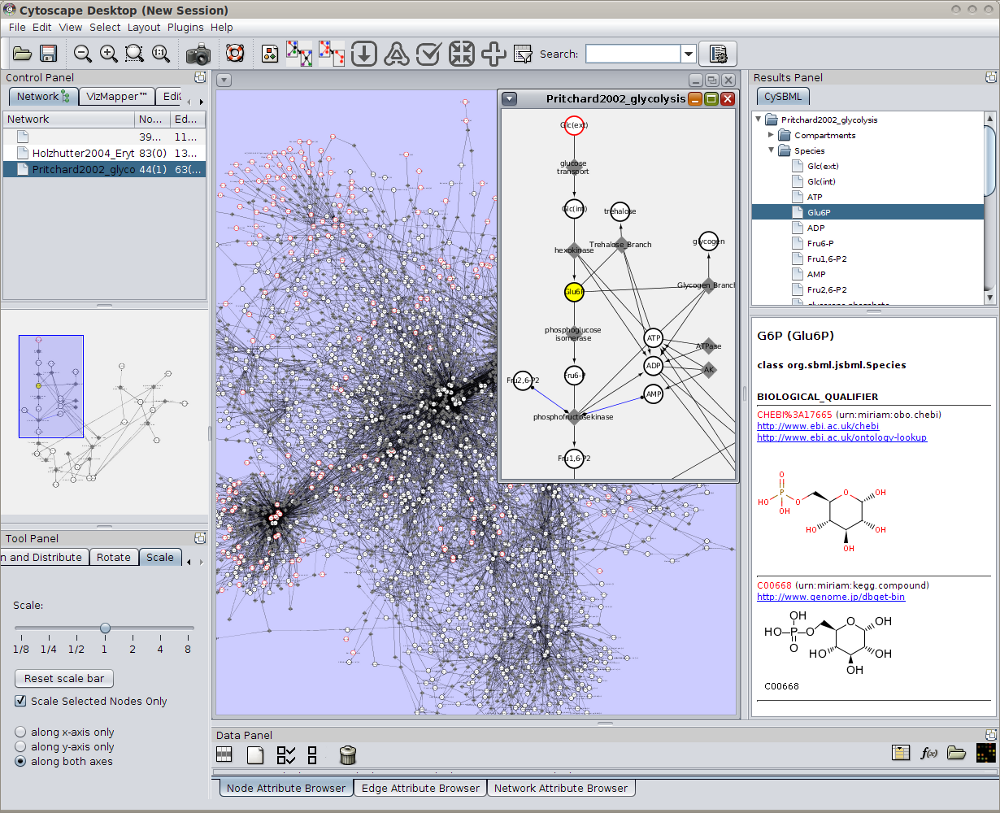
Support of Layout and Qualitative Models Package
cysbml supports the SBML Level 3 packages for qualitative Models and Layouts. Layout information from the layout extension is used to set the positions and boundary boxes of the nodes. Furthermore, SBML qualitative models can be imported with their annotations and thereby allows for models where species do not represent quantity of matter and processes are not reactions per se like, for instance, boolean networks.
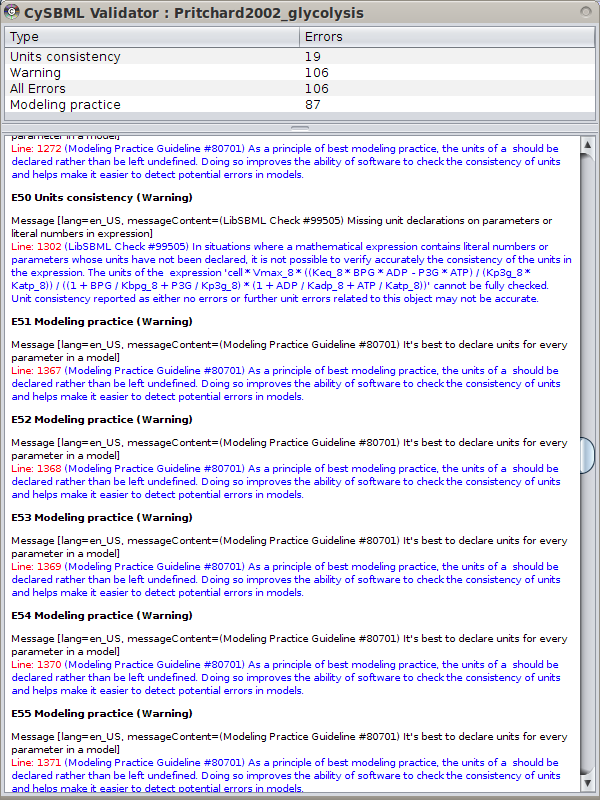
SBML Validation
 SBML models can be validated from within Cytoscape.
Validation errors can be filtered by the severity of the error falling in the classes
SBML models can be validated from within Cytoscape.
Validation errors can be filtered by the severity of the error falling in the classes INFO, WARNING, ERROR, FATAL and ALL.