pymetadata
Python library for COMBINE archives and annotations
Humboldt-Universität zu Berlin, Faculty of Life Science, Institute of Biology, ITB
University of Stuttgart, Institute of Structural Mechanics and Dynamics in Aerospace Engineering
February 2, 2026
COMBINE archive (OMEX)1 ![]()
- *.omex: Consists of a single ZIP-based file (with .omex extension) that bundles models, simulation descriptions, data files, and metadata needed to fully reproduce a study.
- Manifest: Uses a manifest.xml file to catalog contents, including file locations and formats like SBML
- Sharing: Enables sharing complete, reproducible simulation studies rather than scattered files.
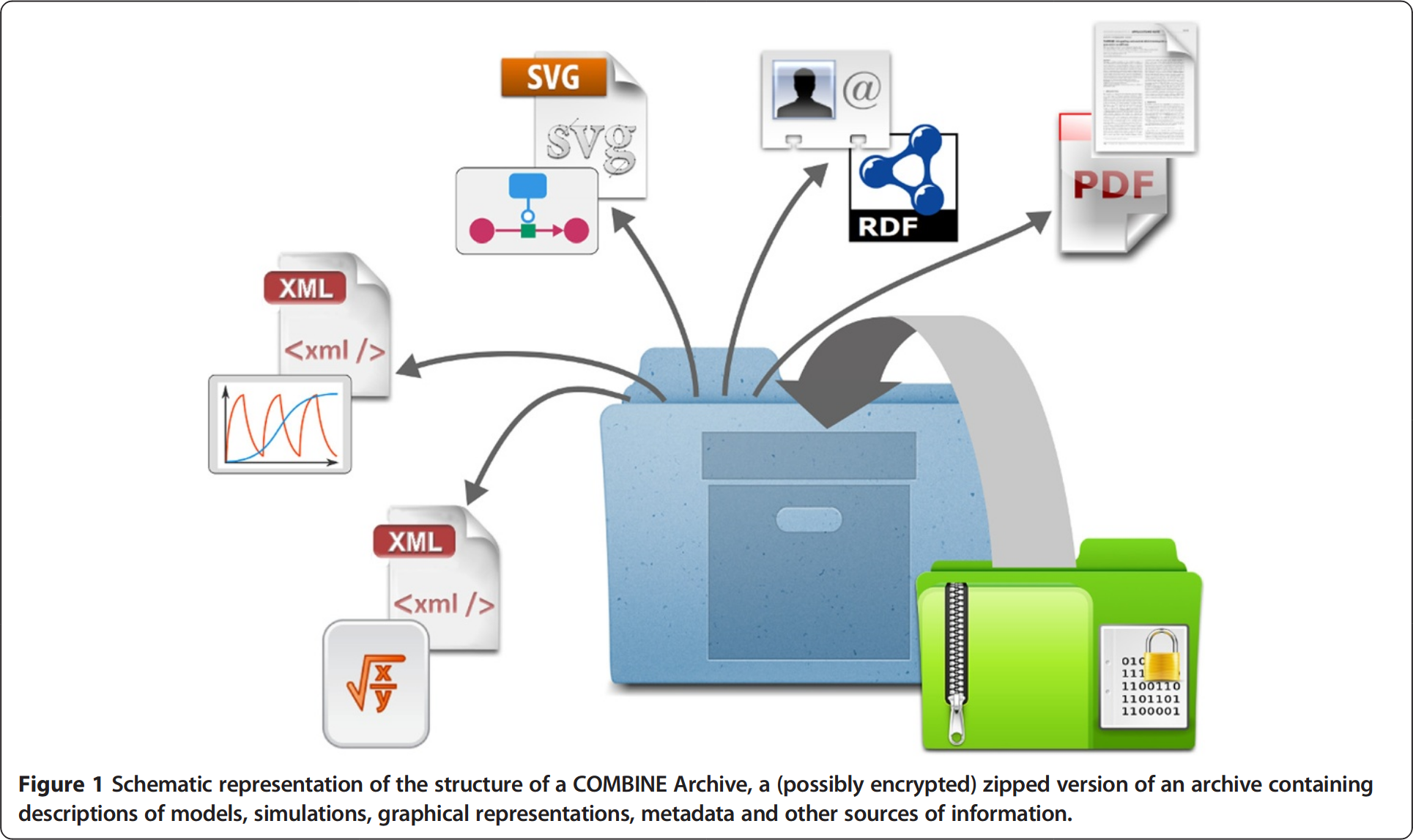
OMEX Metadata
- FAIR_ Metadata is crucial for FAIR1 modelling workflows (findable, interoperable)
- Harmonization: Important to harmonize semantic annotations efforts across standards and modeling domains2
- Description of models, model components and workflows (e.g. canagliflozin-model3)
![]()

pymetadata1
- Supports reading, writing and display of COMBINE archive (OMEX)2
- Validation of metadata against identifiers.org3 and MIRIAM4 registry
- Normalization of metadata
- Resolving metadata via ontology lookup service (OLS4)5
- new: py3.10 - py3.14 support
- new: documentation: https://matthiaskoenig.github.io/pymetadata/
1. Create COMBINE archive
# Create archive from file
omex = Omex()
omex.add_entry(
entry_path=Path("results/testomex/models/omex_comp_flat.xml"),
entry=ManifestEntry(location="./model.xml",
format=EntryFormat.SBML, master=False),
)
omex.add_entry(
entry_path=Path("results/testomex/README.md"),
entry=ManifestEntry(
location="./README.md", format=EntryFormat.MARKDOWN, master=False
),
)
omex.to_omex(Path("results/test_from_files.omex"))
console.print(omex)2. Load from URL
[
ManifestEntry(location='.', format='http://identifiers.org/combine.specifications/omex', master=False),
ManifestEntry(location='./manifest.xml', format='http://identifiers.org/combine.specifications/omex-manifest', master=False),
ManifestEntry(location='./README.md', format='https://purl.org/NET/mediatypes/text/x-markdown', master=False),
ManifestEntry(location='./cc-by-sa-4.0.txt', format='https://purl.org/NET/mediatypes/text/plain', master=False),
ManifestEntry(location='./mit.txt', format='https://purl.org/NET/mediatypes/text/plain', master=False),
ManifestEntry(location='./models/canagliflozin_intestine.xml', format='http://identifiers.org/combine.specifications/sbml.level-3.version-2', master=False),
...
ManifestEntry(location='./figures/canagliflozin_model.png', format='https://purl.org/NET/mediatypes/image/png', master=False)
]3. Working with SBO
- enums for simple access to SBO
4. Harmonizing annotations
from pymetadata.console import console
from pymetadata.identifiers.miriam import BQB, BQM
from pymetadata.core.annotation import RDFAnnotation
for resource in [
"urn:miriam:chebi:CHEBI%3A33699",
"CHEBI:33699",
"chebi/CHEBI:33699",
"https://identifiers.org/chebi/CHEBI:33699",
"http://identifiers.org/CHEBI:33699",
]:
a = RDFAnnotation(qualifier=BQB.IS, resource=resource)
console.print(a)- normalizing multitude of patterns found in models and resources
5. Validating annotations
ERROR MIRIAM namespace `cheb` does not exist for `RDFAnnotation(BQB.IS|cheb|CHEB:33699|identifiers.org)`
RDFAnnotation(BQB.IS|cheb|CHEB:33699|identifiers.org)
ERROR Term `CHEBI:X33699` did not match pattern `^CHEBI:\d+$` for collection `chebi`.
RDFAnnotation(BQB.IS|chebi|CHEBI:X33699|identifiers.org)- validation of namespaces and patterns
Want to know more
Bergmann, Frank T., Richard Adams, Stuart Moodie, Jonathan Cooper, Mihai Glont, Martin Golebiewski, Michael Hucka, et al. 2014. “COMBINE Archive and OMEX Format: One File to Share All Information to Reproduce a Modeling Project.” BMC Bioinformatics 15 (1): 369. https://doi.org/10.1186/s12859-014-0369-z.
Bergmann, Frank T., Nicolas Rodriguez, and Nicolas Le Novère. 2015. “COMBINE Archive Specification Version 1.” Journal of Integrative Bioinformatics 12 (2): 261. https://doi.org/10.2390/biecoll-jib-2015-261.
Bernal-Llinares, Manuel, Javier Ferrer-Gómez, Nick Juty, Carole Goble, Sarala M. Wimalaratne, and Henning Hermjakob. 2021. “Identifiers.org: Compact Identifier Services in the Cloud.” Bioinformatics 37 (12): 1781–82. https://doi.org/10.1093/bioinformatics/btaa864.
König, Matthias. 2026. “Pymetadata Are Python Utilities for Working with Metadata.” Zenodo. https://doi.org/10.5281/zenodo.18207746.
Laibe, Camille, and Nicolas Le Novère. 2007. “MIRIAM Resources: Tools to Generate and Resolve Robust Cross-References in Systems Biology.” BMC Systems Biology 1 (December): 58. https://doi.org/10.1186/1752-0509-1-58.
Neal, Maxwell Lewis, Matthias König, David Nickerson, Göksel Mısırlı, Reza Kalbasi, Andreas Dräger, Koray Atalag, et al. 2019. “Harmonizing Semantic Annotations for Computational Models in Biology.” Briefings in Bioinformatics 20 (2): 540–50. https://doi.org/10.1093/bib/bby087.
Tereshchuk, Vera, Michelle Elias, and Matthias König. 2026a. “A Digital Twin of Canagliflozin Pharmacokinetics and Pharmacodynamics in Type 2 Diabetes Mellitus.” Medicine and Pharmacology. https://doi.org/10.20944/preprints202601.2095.v1.
———. 2026b. “Physiologically Based Pharmacokinetic/ Pharmacodynamic (PBPK/PD) Model of Canagliflozin.” Zenodo. https://doi.org/10.5281/ZENODO.13759839.
Wilkinson, Mark D., Michel Dumontier, I. Jsbrand Jan Aalbersberg, Gabrielle Appleton, Myles Axton, Arie Baak, Niklas Blomberg, et al. 2016. “The FAIR Guiding Principles for Scientific Data Management and Stewardship.” Scientific Data 3 (March): 160018. https://doi.org/10.1038/sdata.2016.18.





